Compound registration
In order to get some compounds into the database you need to register them. You can do that either by filling in the compound registration form and hence add the compounds one-by-one (singleton registration) or you can import the compounds from a file (bulk registration).
Both methods are described below.
Note: before you can register compounds you (or an admin from your organisation) need to create a relevant (matching the compounds you would like to register) compound type.
Singleton registration
To manually register a new compound:
- Click the "Register compound" button
- Select the relevant compound type from the dropdown. (If you miss a compound type you need to add it or ask an admin to do so)
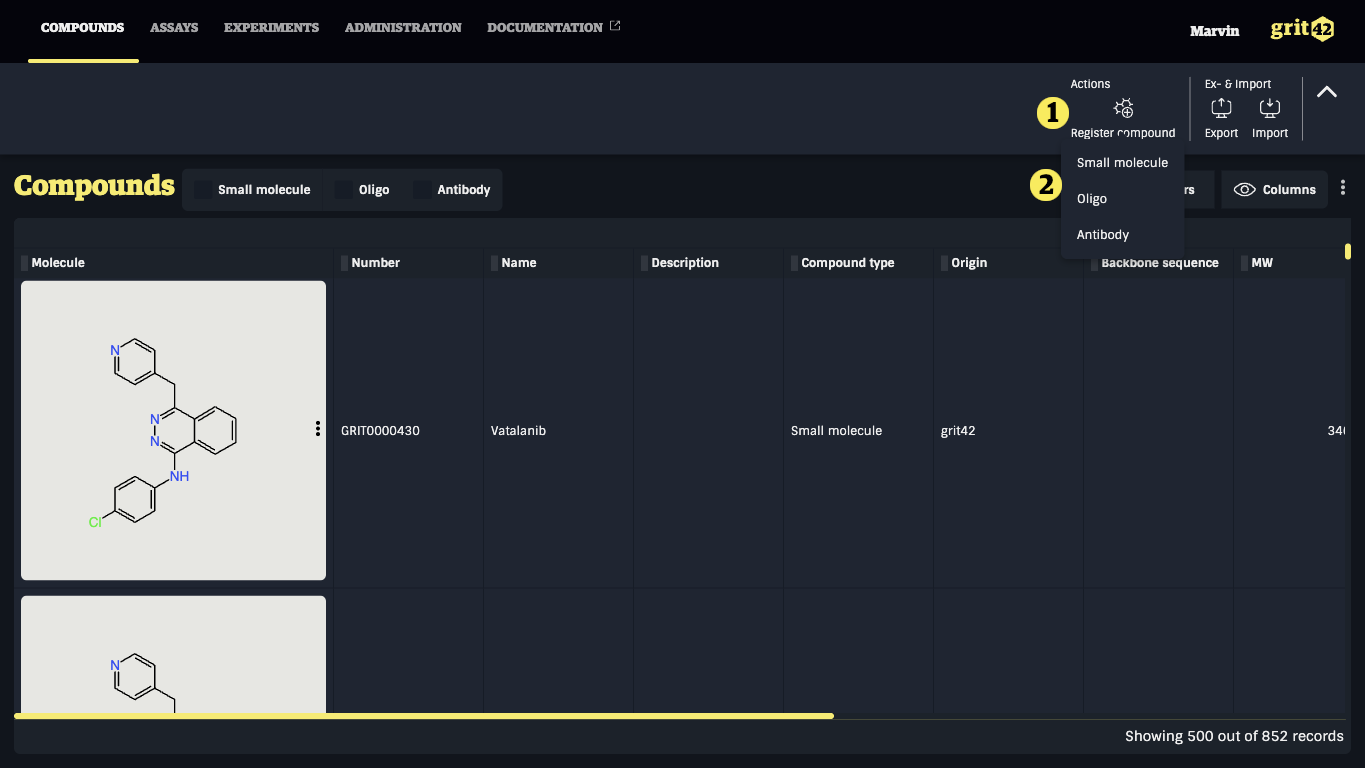
A compound registration form appears (here is shown a small molecule compound type to also illustrate the drawing of the structure)
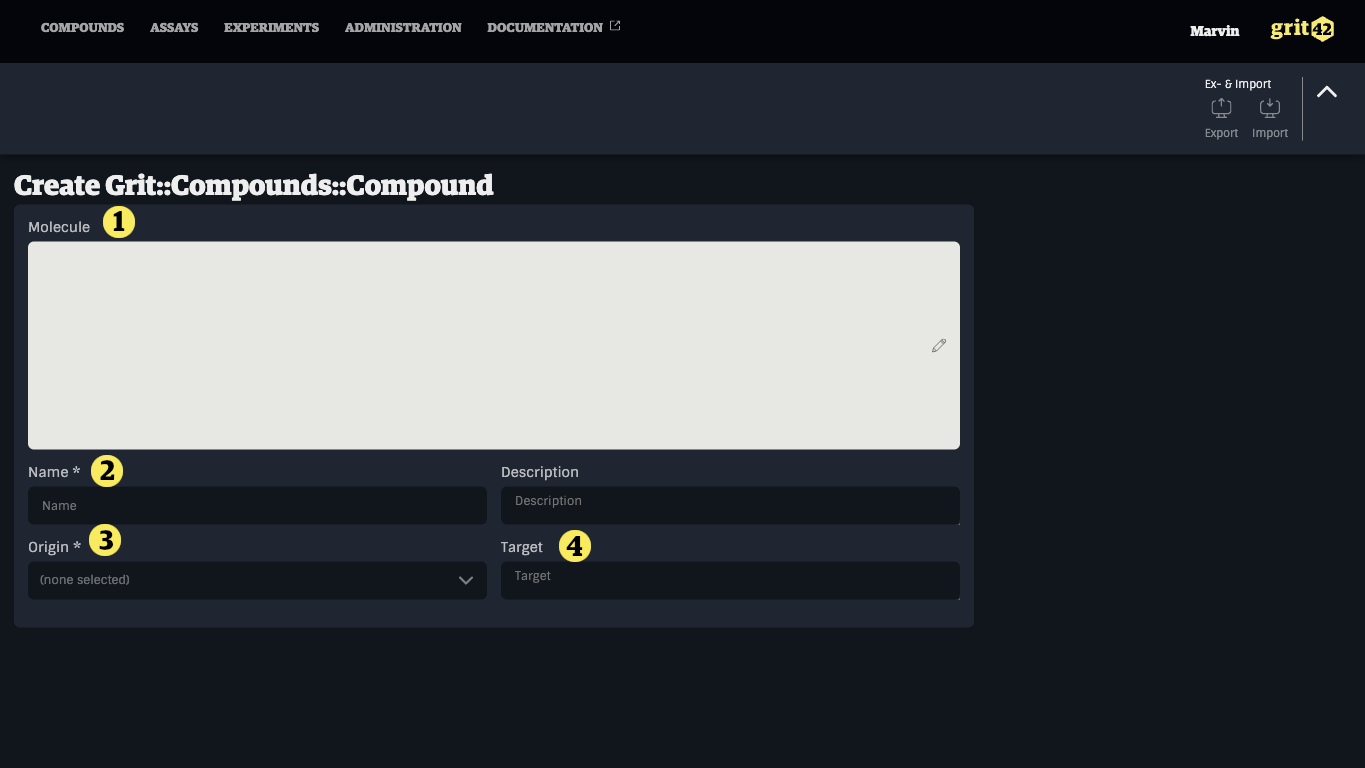
- Field to add a structure drawing (see below)
- Give the compound a name (it will automatically be given a compound ID as part of the process)
- Select the origin from the dropdown
- Add a target name. Here added as an illustration of a specific small molecule compound type property defined for the small molecule compound type.
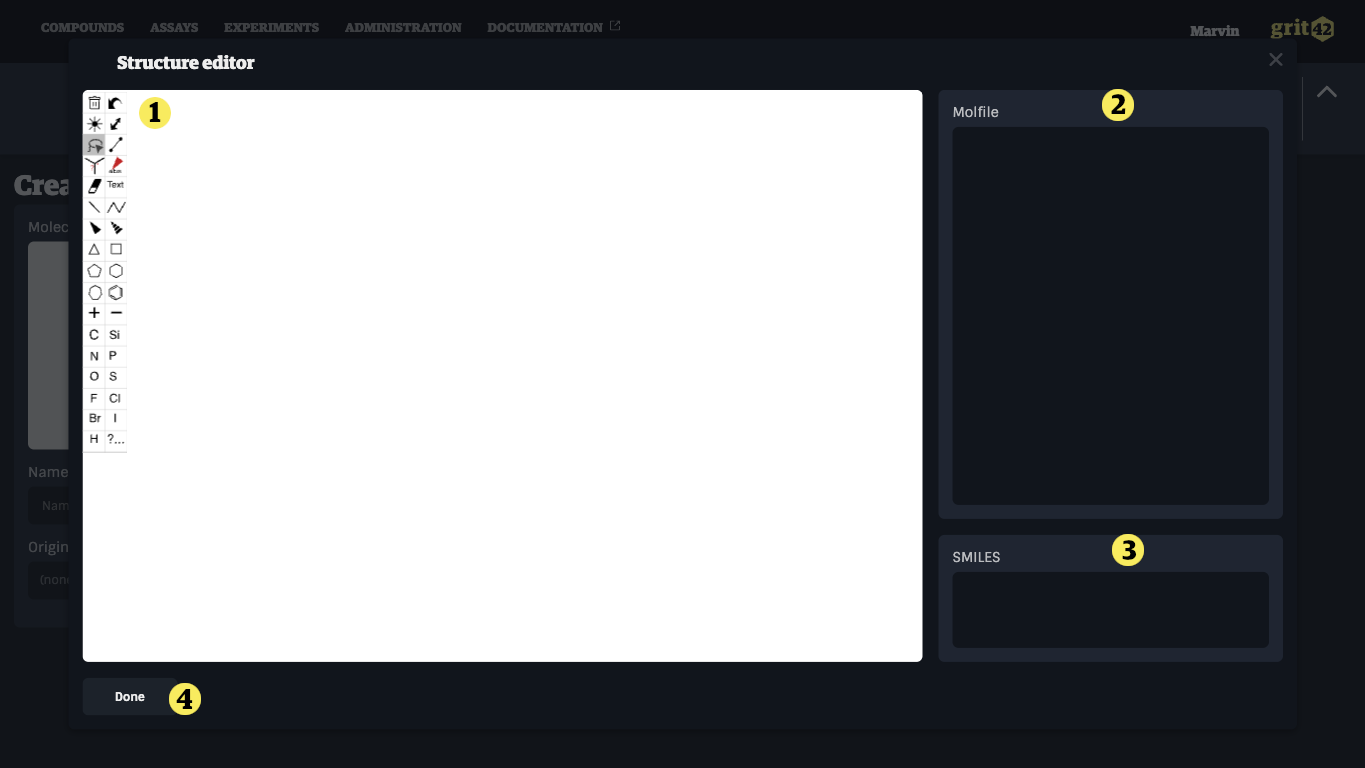
When clicking the molecule field a structure drawing tool appear. (NOTE: IF you want to register a compound without a structure then leave the field empty and go to No structure below)
- Draw the relevant structure using the tools on the left side
- As you draw the molfile will be shown
- As you draw the smiles string will be shown
- When finished click "Done"
The structure will now be added to the compound registration form 1). Fill in the rest of the mandatory fields and click "Save" 2).
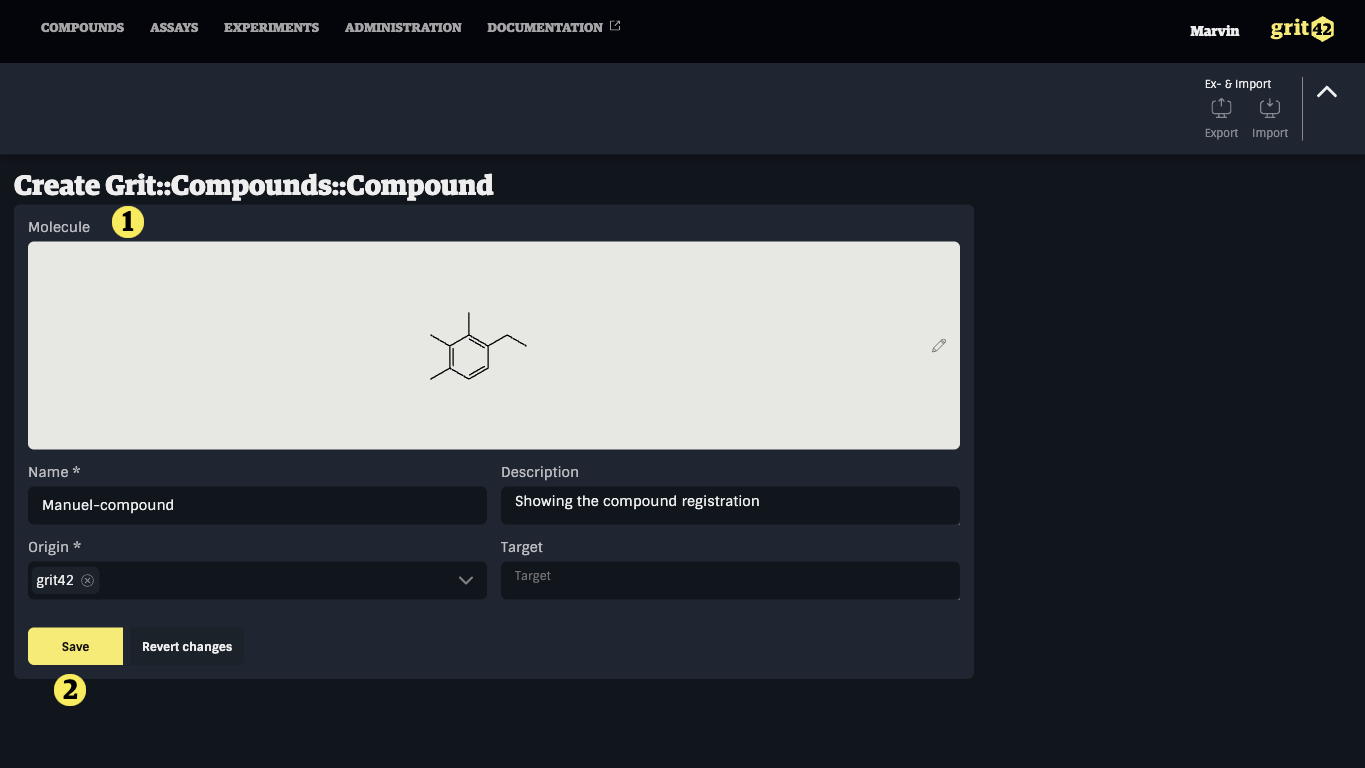
The compound is now registered in the database and the user is taken to the compound details tab.
- The compound number added by the platform as part of the process
- Mw calculated by RDKit on the basis of the structure
- LogP calculated by RDKit on the basis of the structure
- Molecular formula created by RDKit on the basis of the structure
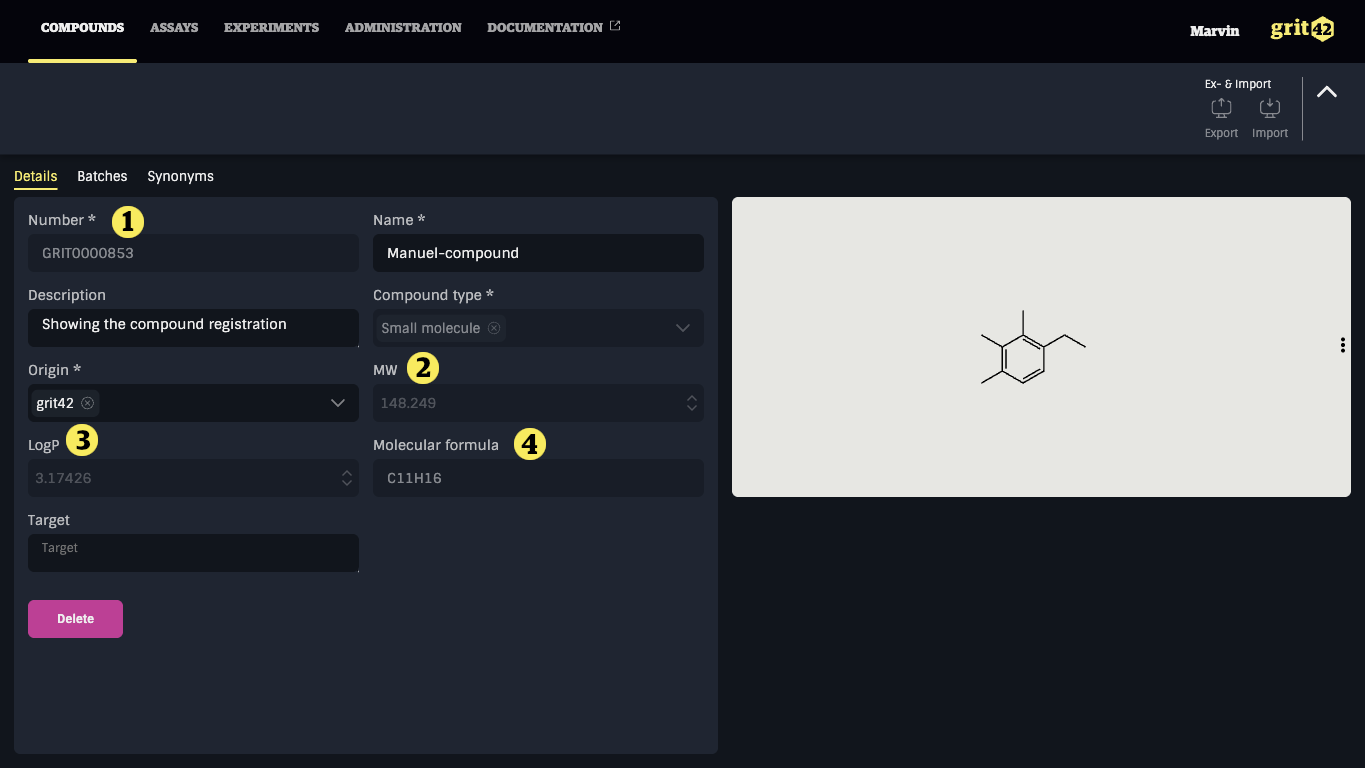
From here the user can delete the compound again or continue to the compounds grid or to add batches under this new compound.
Bulk registration from file
Compounds can be imported from SDF or CSV files. To initiate the compound import process go to the Compounds grid and click the "Import" button 1) and select "import compounds" in the appearing dropdown.
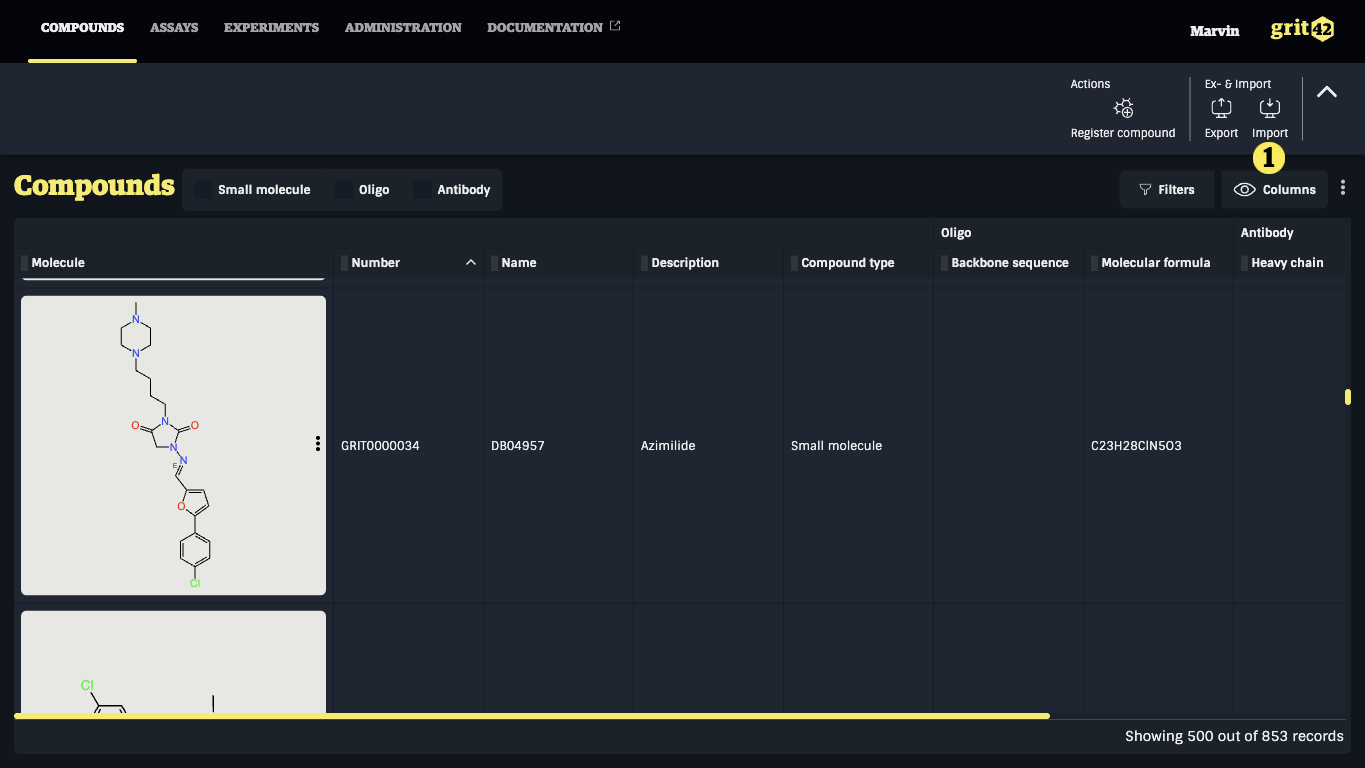
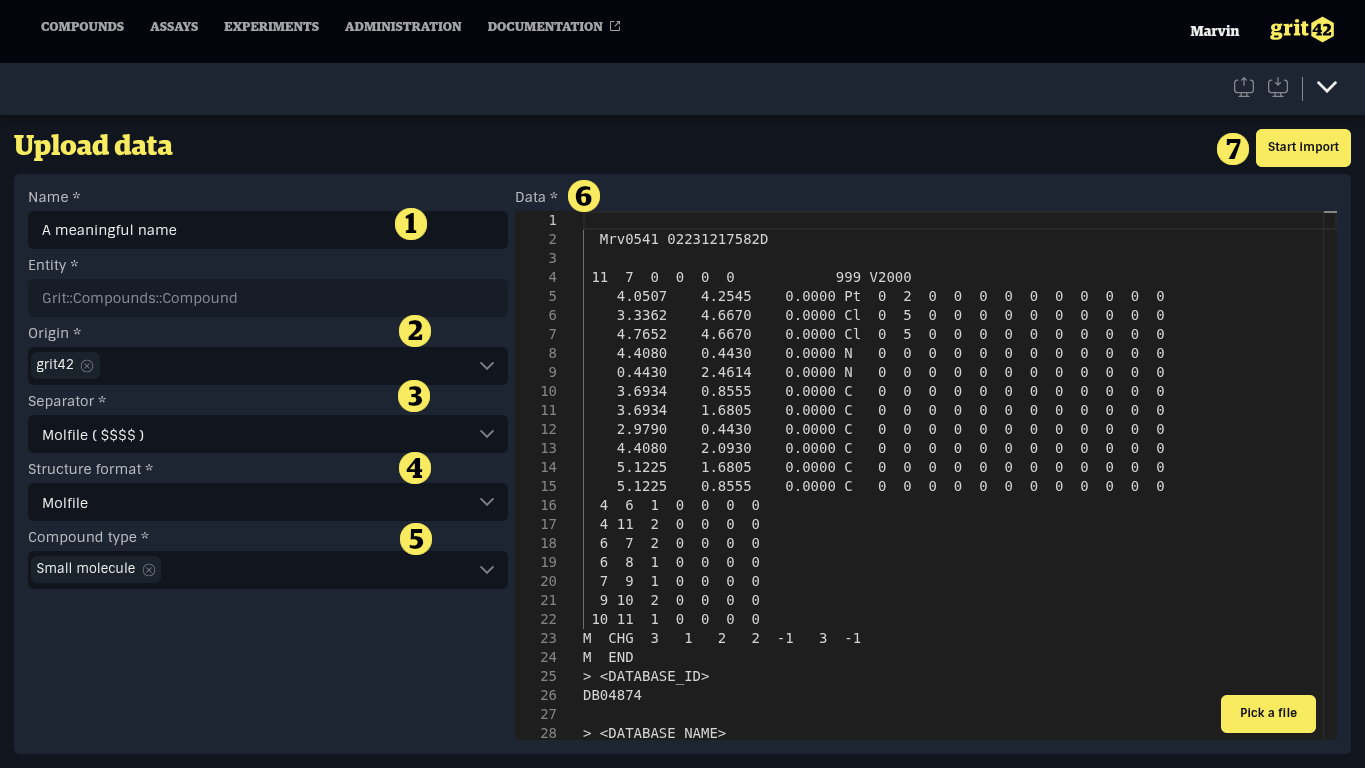
This will take you to the import form:
- The name of the load. The platform creates an automatic name, but you can edit. The name can be used to identify this particular load later
- Select the relevant origin of the compounds from the dropdown
- The separator used in the data set. When adding a file, the system will attempt to guess the separator used based on the content of the file
- The format of structures in the data set. Supported formats are Molfile in SDF data sets and SMILES in CSV data sets. When adding a file, the system will attempt to guess the structure format based on the content of the file.
- The compound type. The selected compound type will determine the mappings available in the next step.
- The data set. A data set can be added by:
- dropping a file in the dark area
- opening the file explorer and selecting a file
- clicking the dark area and typing manually or pasting a selection from a spreadsheet or another text editor
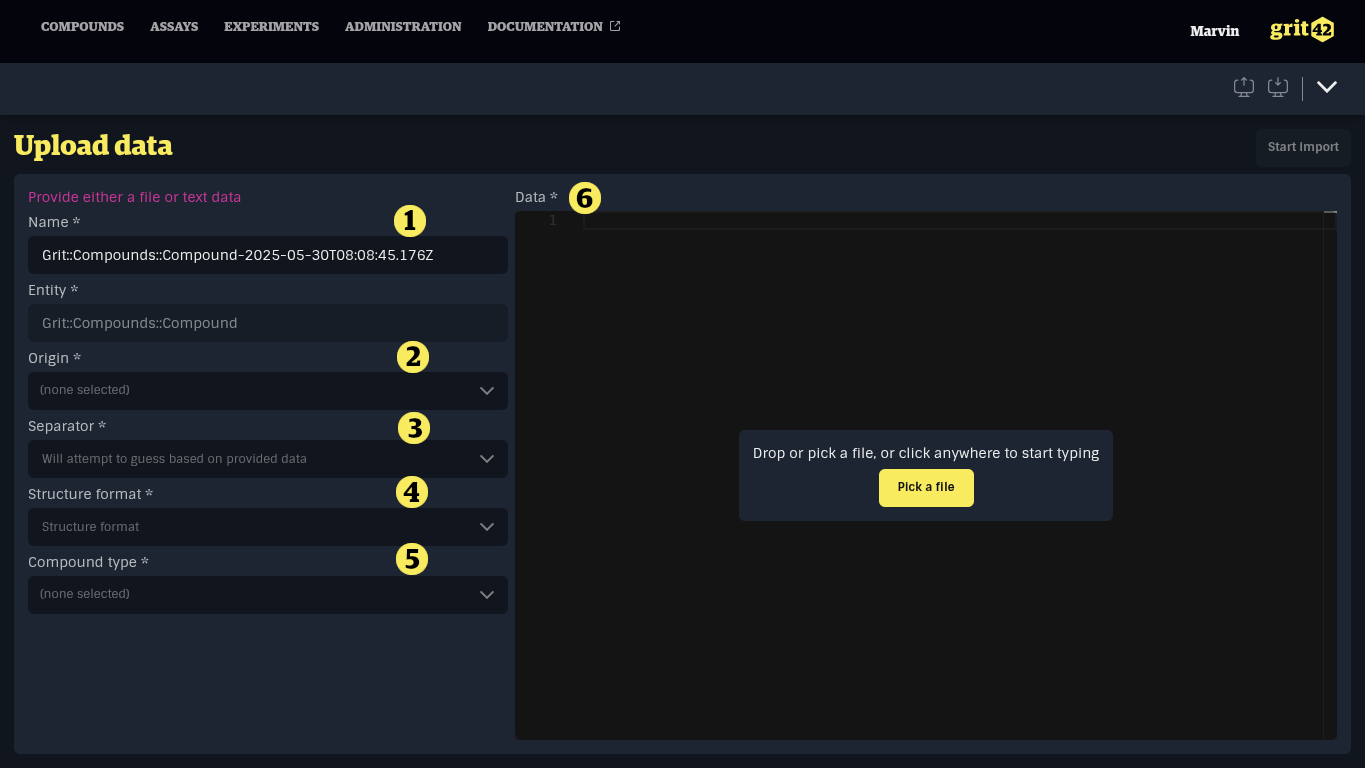
When the form has been filled in it can look like this.
Go to the button of the screen and click "Start import" (7)
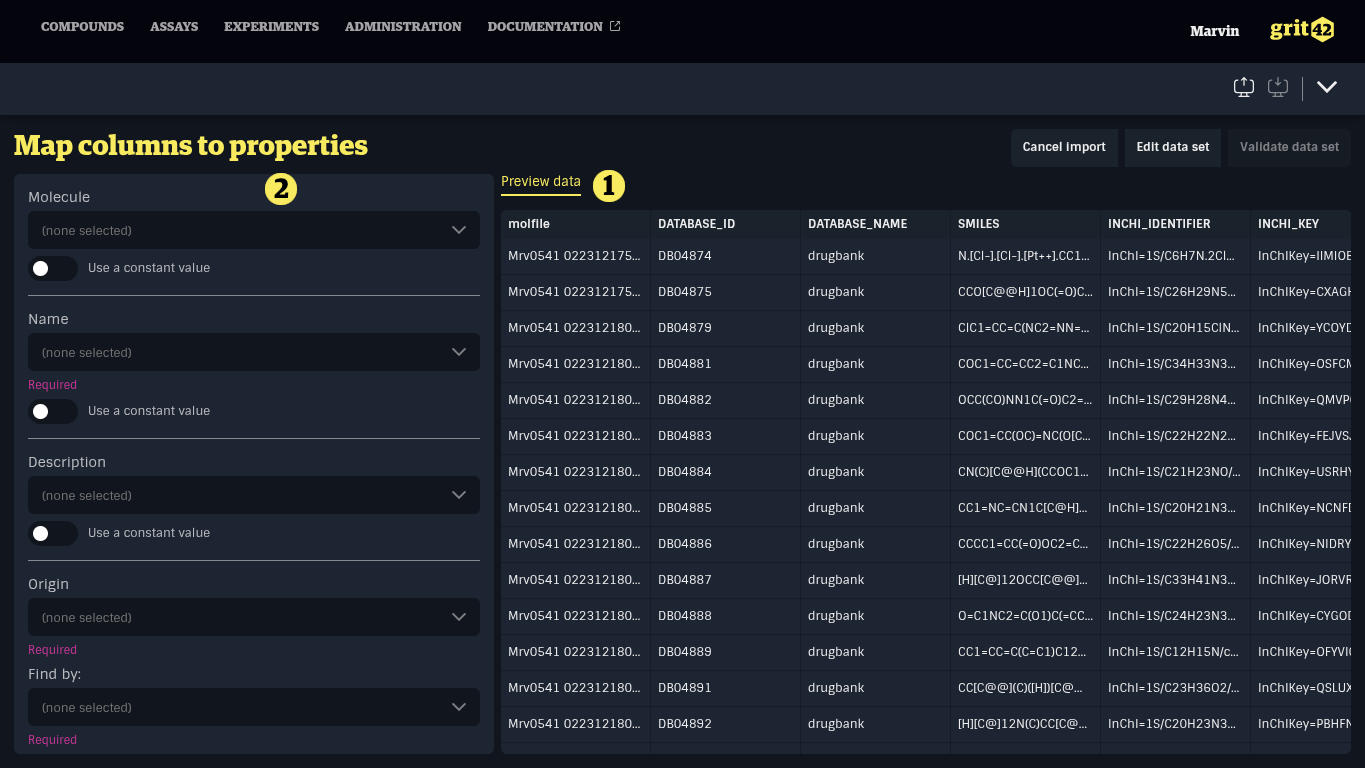
The next step is the mapping.
Here you map the relevant columns from the file 1) into the matching columns in the database 2)
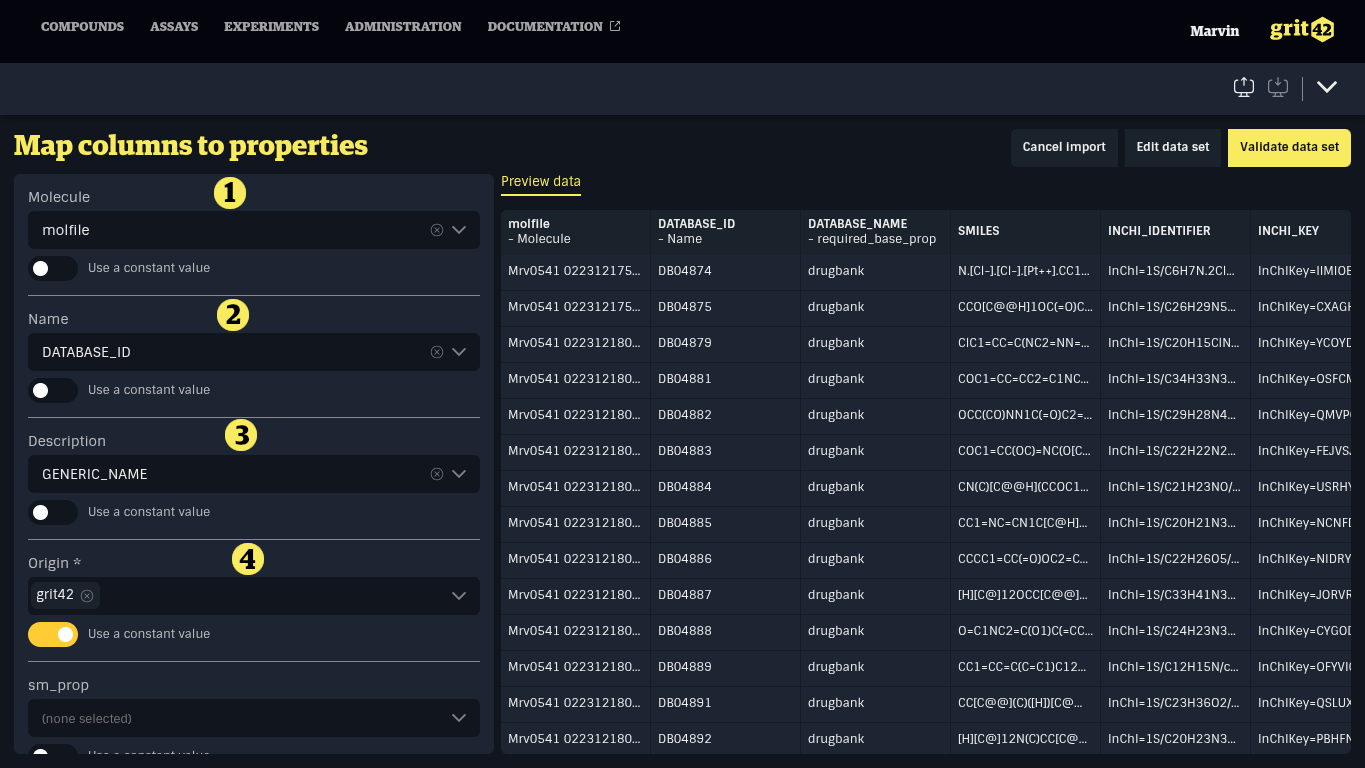
The mapping can look something like this:
- The molfile field from the file is mapped to the Molecule field in the database
- The DRUGBANK ID from the file is mapped to the Name of the compound in the database
- The GENERIC_NAME from the file will be added to the Description field
- As the file does not contain a relevant origin of the compounds we have here turned the switch "Use a constant value" on and selected a value from the platform list of origins. The origin (here grit42) will be added to all the compounds when they are loaded
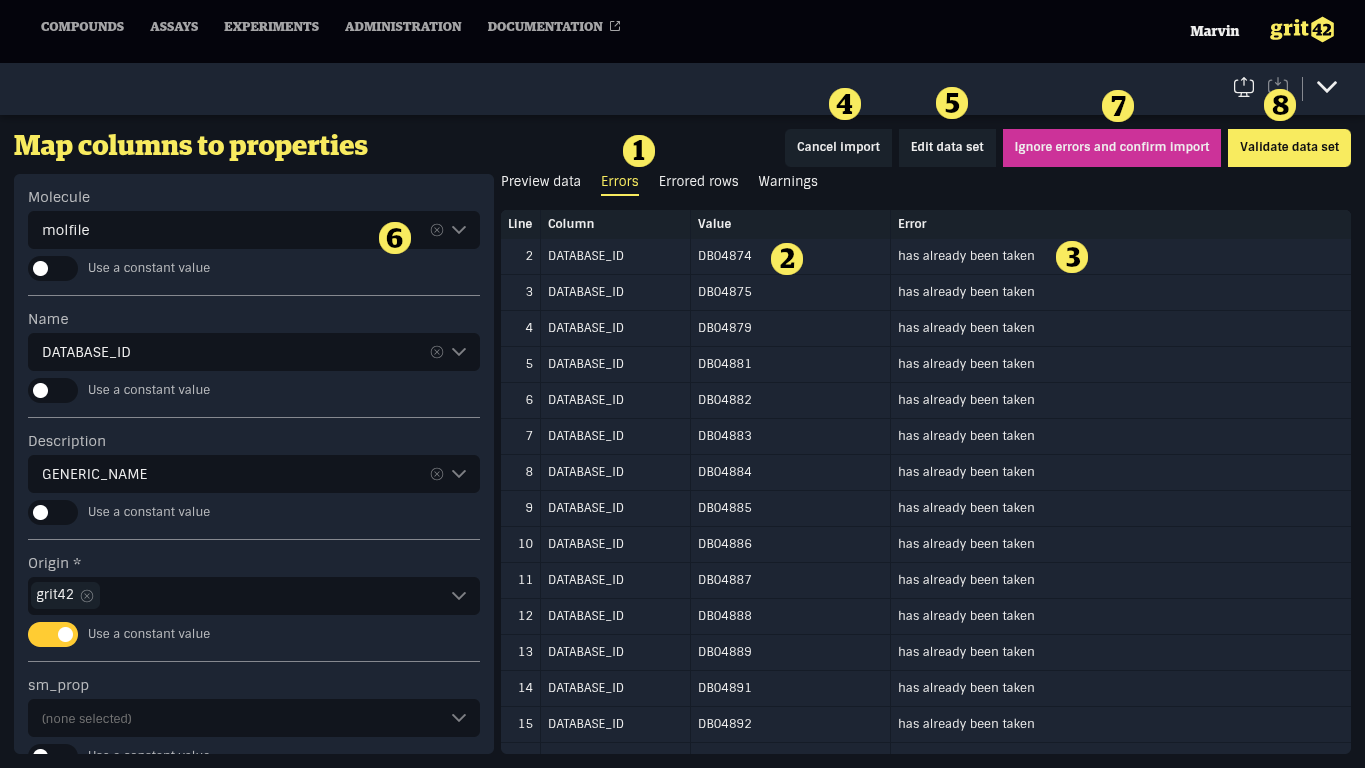
But in this case the import was aborted by the platform.
Three new tabs (Errors, Errored rows, Warnings) 1) appear next to the file data preview and errors are shown. The data show in these tabs can be exported for further processing.
In this case the issue is that the value (the DRUGBANK ID we mapped to the name column) 2) must be unique in the database (so we only have one compound with that name) but as these compounds have been loaded before 3) these DRUGBANK ID's already exist.
Resolution options are:
- Change the mapping of the Name column to another relevant name/ID in the file if that exist (most compounds have an ID from the producer)
- Cancel the import now and start over (4)
- Edit the data set manually by selecting a new file or manually editing the loaded data set (5)
- Ignore the rows producing errors and load only valid records (6)
- Validate the data set again, for instance after modifying existing compounds (7)
If there are no issues, the user will be asked to confirm the import.
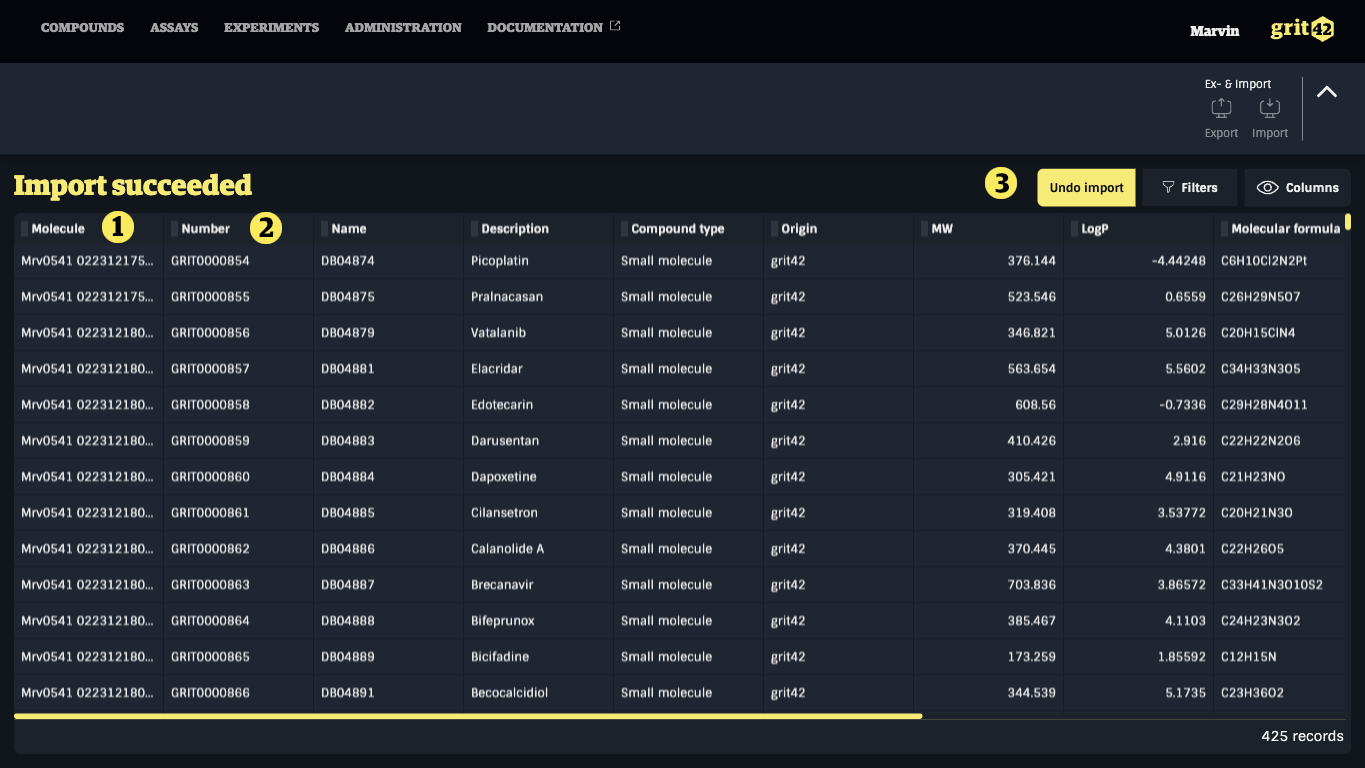
Once the import is confirmed or only valid records are loaded, the system will display an 'Import Succeeded' view, indicating that the data set has been successfully added to the database.
- The structures from the file are loaded into the Molecule field
- The compounds have automatically been given a grit database ID
- The user can here decide to undo the import whereby the whole loadset will be retrieved again
No structure - registering a new compound without any structure/molecule
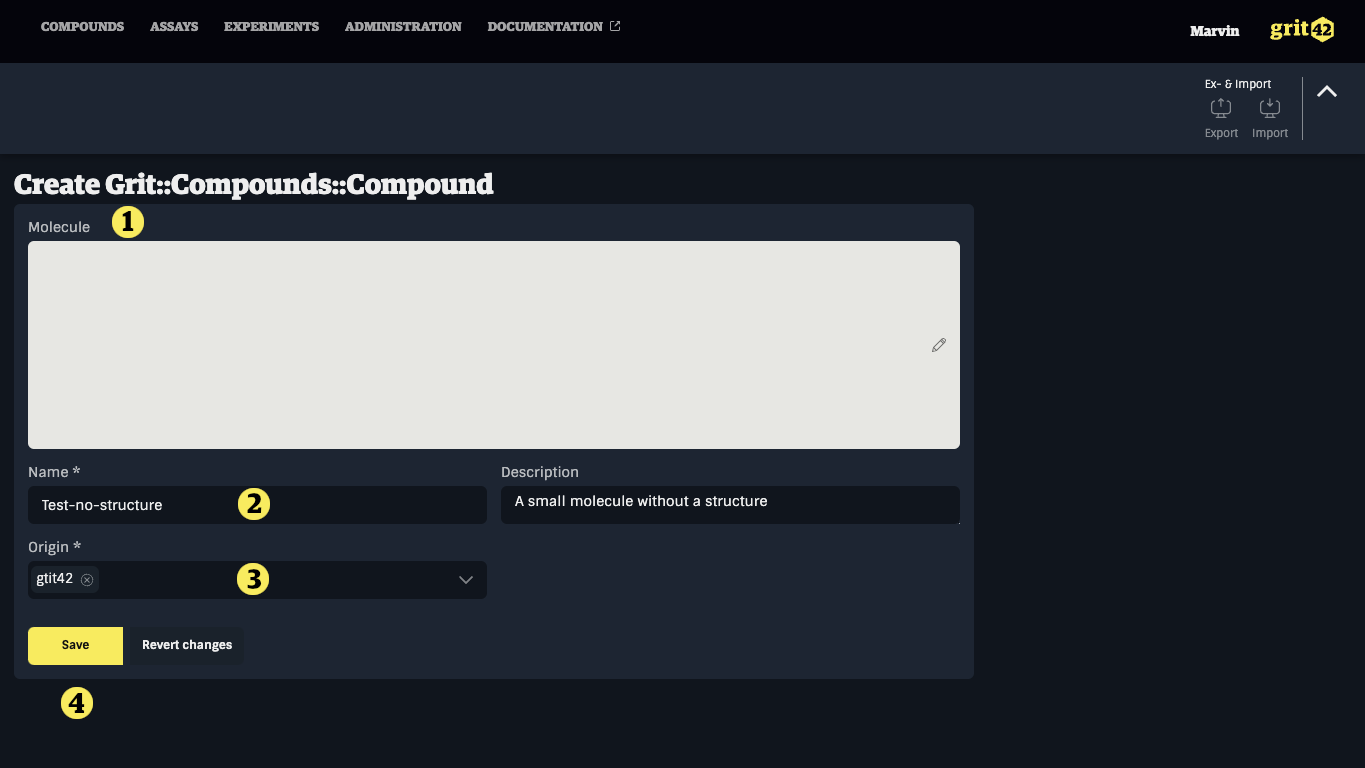
To manually register a new compound without a structure:
- Click the "Register compound" button
- Select the relevant compound type from the dropdown. (If you miss a compound type you need to add it or ask an admin to do so)

A compound registration form appears:
- Field to add a structure drawing. As we are going to register a compound without a structure skip this field and continue
- Give the compound a name (it will automatically be given a compound ID as part of the process)
- Select the origin from the dropdown
- Click save
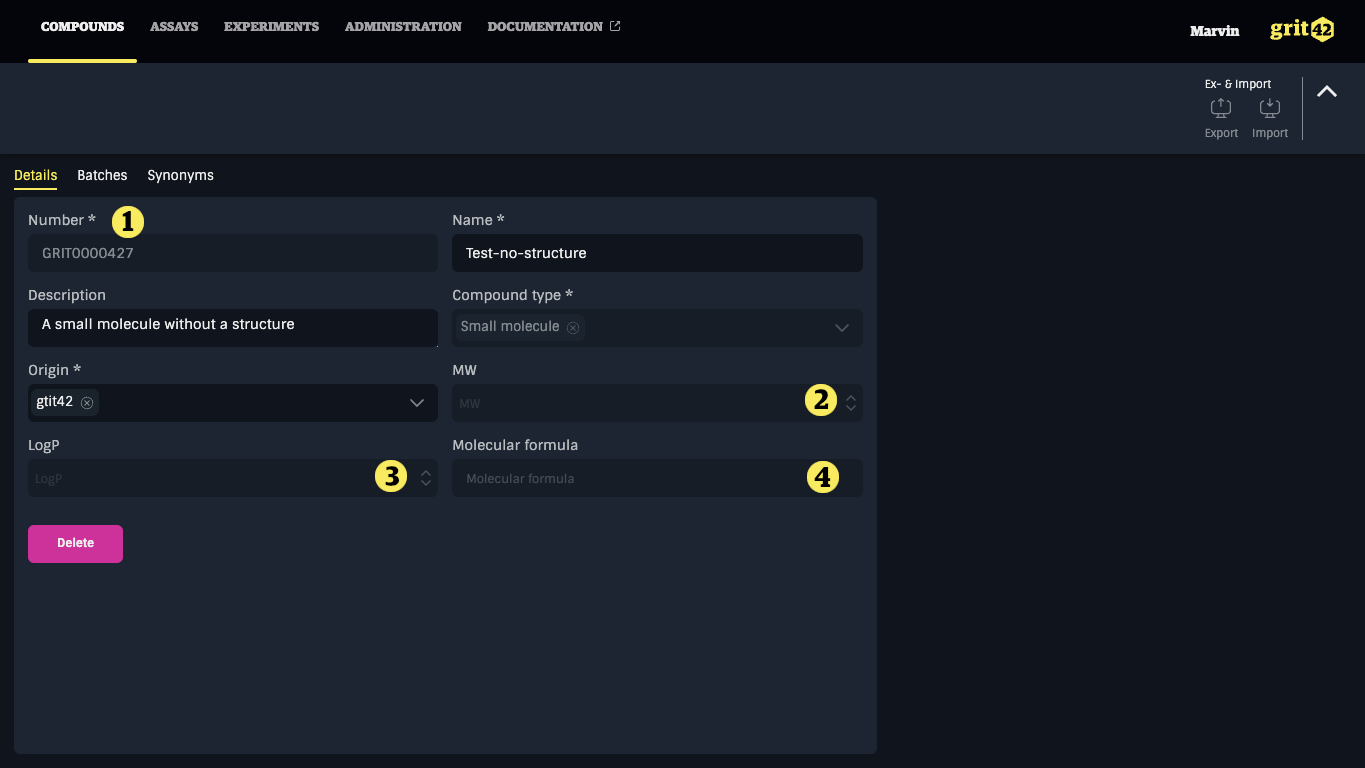
The compound has now been registered in the database and been assigned a compound number 1).
As the compound doesn't have a structure attached to it the automatically calculated properties (2, 3 and 4) will be empty.
The compound can be seen in the Compounds grid with the assigned compound number 1). The structure (Molecule) field is naturally empty 2).